

| HOME | SERVICE | PRODUCT | SUPPORT | ORDERING | CONTACT |
Got Mold?
Analysis via DNA Quantification
By Steve Vesper, Ph.D. & King-Teh Lin, Ph.D.
Got Mold? The question has become more and more relevant to many industries, but trying to answer this question can be difficult.
Traditionally, mold analysis has been performed by either microscopic observation or plate culturing of molds on various media. These laborious techniques were developed in the 19th century and lack the standardization we expect in today's technologically advanced world. The analytical laboratory of the 2lst century offers a solution to mold analysis arising from powerful technological advances in DNA detection, notably a method called Quantitative Polymerase Chain Reaction (QPCR).
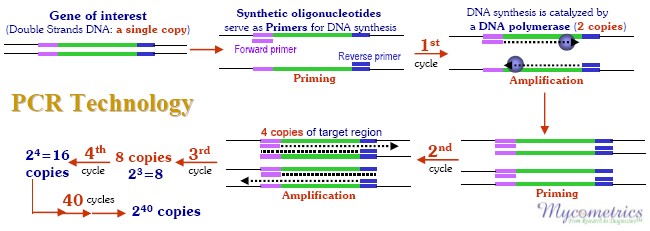
In 1993 the Nobel Prize was awarded1 for the development of a method to make many copies of DNA sequences clipped from their original genomes. By using the enzyme DNA polymerase, a large number of copies of isolated DNA sequences can be made in a test tube to enable scientists to study these sequences with ease. DNA sequences are the strings of nucleotides containing the four distinguishing identifiers: A, C, G and T.
Sequential combinations of these four "bases" form the genomic blueprint ("genes") responsible for making each creature, including different mold species, unique. The process of copying DNA sequences by amplification with DNA polymerase has become known as the Polymerase Chain Reaction (PCR)2.
For PCR, short, very specific sequences of DNA called the forward and reverse primers are selected from the DNA sequence of the target mold genome. These primers are what allow the PCR process to make copies of or "amplify" the isolated target DNA (see figure below). As PCR amplification proceeds, copies of the target DNA sequences accumulate. The number of these accumulated copies can be estimated but not very accurately. PCR technology, as practiced before 1997, had great ability to multiply the specific isolated DNA sequences, but it lacked good quantitation, thus limiting its use in many applications.
In 1997 Applied Biosystems, Inc. announced the release of a new instrument called the Sequence Detector, which provided a very accurate way to count the accumulated PCR copies. This measurement was accomplished by adding a fluorescently labeled species-specific DNA sequence, called the "probe," into the PCR amplification. This new process now goes by the name Quantitative PCR or QPCR.
The QPCR Process
QPCR analysis begins by extracting the DNA from the mold cells in a sample. The sample could be dust from a house, flour from a mill where the level of aflatoxin producing molds is critical, collected particles from a hospital during nearby construction, bulk samples from water-damaged materials or water from a bottling plant.
The samples are processed in a matter of minutes using a standard protocol, and the DNA is ready for QPCR analysis. QPCR detects the individual mold species present, and furthermore, yields a quantitative report on the presence of each mold.
Primers and probes for the QPCR process are the key, and these must be species-specific. The work on mold-specific QPCR has advanced to the point that there are now over 100 sets of primers and probes available for specific molds3.
A mold-specific QPCR reaction mixture contains the primers and probes for a specific mold species together with the amplification solution. To this mixture is added a small aliquot of the sample DNA extract. This process is repeated for each of the target specific mold species, each ready to detect the presence of a specific mold. The reaction tubes are then placed in the Sequence Detector instrument. Here the primers and probe will bind or "hybridize" to the one mold DNA they recognize within the DNA extract. When there is an exact DNA match, fluorescence from the probe is released, and the Sequence Detector tallies the fluorescence from each reaction mixture.
Concurrently, this same analysis is performed on a "standard" spore suspension which contains a known number of spores of a specific target mold. The standard functions as the internal calibration for the process. By comparing the fluorescence from the standard spore suspension extract to the fluorescence from the environmental sample extract, the analyst can determine the number of target mold spores in the environmental sample. Through this process, quantitative analysis can be accomplished for as many specific environmental molds as desired.
Advantages of QPCR
QPCR offers many advantages to industrial hygienists, environmental consultants, indoor air quality professionals, building owners, home inspectors, businesses, hospitals, clinics, public/private health agencies and professionals in many other industries.
QPCR will accurately and precisely detect all the molds contained in the sample if the species-specific primers and probes are provided. Those molds that are not detected by traditional culturing techniques can be easily quantified by QPCR.
Applications of QPCR
QPCR can be used for many different applications and projects. In a recently published article4, QPCR was applied to monitor the decontamination of mold during the construction process of a hospital addition where significant water intrusion had occurred. The on-time completion of the construction and a final report certifying that the area was clear of any contamination were required for this project.
QPCR was selected due to its rapid turnaround time, sensitive detection and more representative air sampling. Using QPCR, the engineers rapidly pin-pointed the problem to the correct locations and resolved the mold problem quickly. The QPCR data also facilitated the discovery of other contaminated sources and led to their removal before construction continued. The QPCR results clearly revealed that the cleaning process was effective and that the areas in question were free of fungal contamination.
A similar kind of analysis was performed at a hospital during a renovation process that involved old carpet removal. Since carpet is a well known reservoir of indoor molds, hospital administrators were most interested in protecting the seriously ill burn patients from potential secondary infections arising from aerosolized molds during the renovation work5. They too found the quantitation, specificity and rapidity of QPCR were essential in helping them track mold presence during carpet removal and the subsequent clean-up processes.
QPCR can be a cost effective way to facilitate the evaluation and remediation process. For example, one could select a few dominant mold species to use as a clearance indicator after screening the mold species from the contaminated sites by a combination of culturing techniques and QPCR. Because of the advantages of QPCR, a consulting firm working on a construction project could combine both culture techniques and QPCR to identify certain signature mold species. With the selection of a few representative signature mold species as clearance indicators, the firm could continue remediation in a cost effective and timely manner.
Another intriguing application of QPCR was developed6 in response to the increasing concern over structural damage to buildings caused by wood decay fungi (also referred to as brown rot fungi). These fungi typically do not develop the morphological characteristics for phenotypic identification under laboratory conditions. Therefore, it is difficult to provide a proper diagnosis of early decay or to arrest decay prior to loss of structural integrity. However, QPCR uses genetic identification to provide a rapid and reliable procedure for the early diagnosis of fungal infestation. For example, a residential building was suspected to be infested with a brown rot wood decay fungus, although its structure appeared sound. Using QPCR, it was possible to provide a positive identification and confirmation of the causative agents of the decay so that appropriate remediation could be started.
QPCR can also be used to monitor bacteria like Legionella pneumophila, the cause of Legionaire’s disease. In a recent study submitted for publication7, we were able to monitor the water in the homes of Legionella pneumonia victims to determine if the home might have been the source of the bacterium. The technology is the same; only the primers and probes differ.
QPCR is available today for applications from indoor or outdoor environmental analysis (air, dust, bulk, swabs, liquid etc), food testing, monitoring drug manufacturing, clearing construction projects, measuring molds in soil or agricultural products and even testing for Legionella and other bacteria in the air or water.
NOTICE
The U.S. Environmental Protection Agency (EPA) through its Office of Research and Development, partially funded and collaborated in the research described here. It has been subjected to the Agency's peer review and has been approved as an EPA publication. Mention of trade names or commercial products does not constitute endorsement or recommendation by the EPA for use.
About the Authors
Steve Vesper, Ph.D., is an environmental microbiologist with the U.S. EPA, vesper.stephen@epa.gov, US Environmental Protection Agency, 26 W. M. L. King Drive, Cincinnati, Ohio 45268.
Dr. Vesper received his Ph.D. in Environmental Microbiology from Ohio State University in 1983. He was microbiologist at Battelle Memorial Institute for four years before becoming head of Microbiology at Henkel-Emery Corporation where he obtained a series of patents on new enzymes for natural product applications. From 1990 to 1998, he was a Research Professor in the Department of Civil and Environmental Engineering at the University of Cincinnati where he developed and patented new remediation technologies. In 1998, he joined the US EPA National Exposure Research Laboratory and has published 25 papers and book chapters in the last five years.
King-Teh Lin, Ph.D., is Laboratory Director at Mycometrics, LLC, Monmouth Junction, NJ 08852, kingteh@mycometrics.com.
Dr. King-Teh Lin earned his Ph.D. from the Department of Molecular Genetics, Microbiology and Immunology at Robert Wood Johnson Medical School, UMDNJ in 1997. After earning his degree and completing his postdoctoral fellowship, he continued on at the same University as an instructor, up until 2001. Dr. Lin went on to join P&K Microbiology Services as a Director of Research and Development, where he commercialized the EPA-licensed mold QPCR technology and invented a new method for identification of wood decay fungi. His work has been published in many leading peer-reviewed Journals. In 2005, he established Mycometrics, LLC. Dr. Lin is currently Laboratory Director at Mycometrics, and is an Adjunct Instructor at UMDNJ.
References
1. http://nobelprize.org/chemistry/laureates/1993/press.html.
2. Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, and Arnheim N., 1985 Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 230 (4732):1350-4.
3. http://www.epa.gov/microbes/moldtech.htm.
4. Morrison, J., Yang, C., Lin, K.-T., Haugland, R.A., Neely, A. N., and Vesper, S. J. 2004 Monitoring Aspergillus species by quantitative PCR during construction of a multi-story hospital building. Journal of Hospital Infection. 57:85-87.
5. Neely, A. N., Gallardo, V., Barth, E, Haugland, R. A., Warden, G., and Vesper, S. J. 2004. Rapid monitoring by QPCR for pathogenic Aspergillus during carpet removal from a hospital. Infection Control and Hospital Epidemiology. 25:350-352.
6. Lin, K. T., Li, D. W., Dennis, D. A., Woodcock, R., and Yang, C. S. 2005. Qualitative identification of Meruliporia incrassata using real time polymerase chain reaction (PCR). In Bioaerosols, Fungi, Bacteria, Mycotoxins and Human Health: Patho-physiology, clinical effects, exposure assessment, prevention and control in indoor environments and work. E. Johanning ed., Fungal Research Group Foundation, New York, PP 335-342.
7. Vesper, S. J., Rogers, M. E., Wymer, L. J., Dufour A, and Haugland, R. A. 2006. Opportunistic Pathogens Legionella and Aspergillus Occurrence in Water Supply of Homes of Legionellosis Positive Patients. Submitted to: Epidemiology and Infection.
This article is published by Mycometrics, LLC. For additional information, references and assistances, contact Mycometrics LLC by fax at 732-658-5185, by telephone at 732-355-9018, or email: quest@mycometrics.com.
©All rights reserved. No part of this work may be reproduced or used in any form or by any means - graphic, electronic, photocopying, recording or taping - without the written consent of Mycometrics, LLC. Published date: 9/28/2005
| Copyright © 2005-2022 Mycometrics, LLC. All rights reserved. |